Population structure of community-acquired extended-spectrum beta-lactamase producing Escherichia coli and methicillin resistant Staphylococcus aureus [...]
ABSTRACT
BACKGROUND
Antimicrobial resistance is a global health issue and extended-spectrum β-lactamase producing Escherichia coli (ESBL-Ec) and methicillin-resistant Staphylococcus aureus (MRSA) are of particular concern. Whole genome sequencing analysis of isolates from the community is essential to understand the circulation of those multidrug-resistant bacteria.
OBJECTIVES
Our main objective was to determine the population structure of clinical ESBL-Ec and MRSA isolated in the community setting of a French region.
METHODS
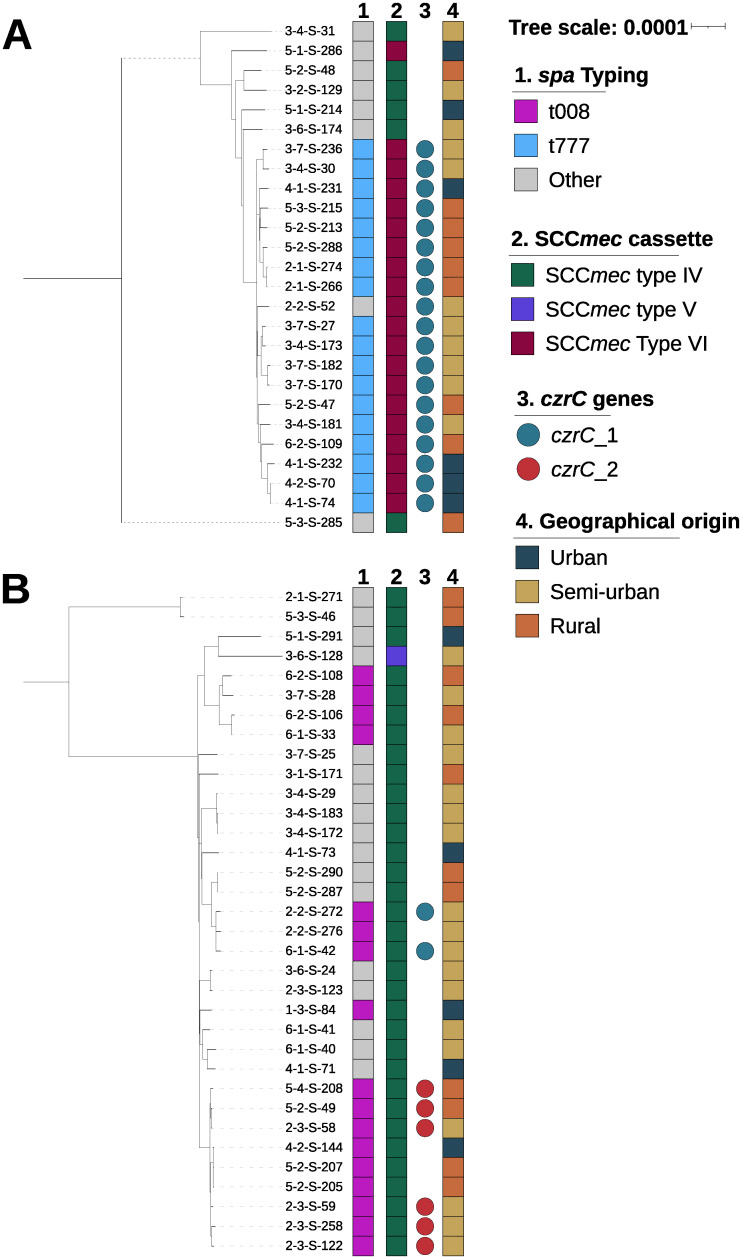
For this purpose, isolates were collected from 23 sites belonging to 6 private medical biology laboratories in the Bourgogne-Franche-Comté region. One hundred ninety ESBL-Ec and 67 MRSA were sequenced using the Illumina technology. Genomic analyses were performed to determine the bacterial typing, presence of antibiotic resistance genes, metal resistance genes as well as virulence genes.
RESULTS
Analysis showed that ST131 was the major ESBL-Ec clone circulating in the region, representing 42.1% of the ESBL-Ec isolates. The blaCTX-M genes represented 98% of blaESBL with the majority being blaCTX-M-15 (53.9%). MRSA population consisted of mainly of CC8 (50.7%) and CC5 (38.8%) clonal complexes. Interestingly, we found a prevalence of 40% of the zinc resistance gene czrC in our MRSA population.
CONCLUSION
We observed no differences in our ESBL-Ec or MRSA populations between urban and rural areas in our French region, suggesting no impact of population density or rural environment.