Genomic epidemiology of third-generation cephalosporin-resistant Escherichia coli from companion animals and human infections in Europe
ABSTRACT
BACKGROUND
In high-income countries, dogs and cats are often considered members of the family. Because of this proximity, it has been suggested that pets and humans might exchange bacterial species from their gut microbiota, with multidrug resistant bacteria being of particular concern.
OBJECTIVES
The aim of this study was to compare the genomes of third-generation cephalosporin-resistant (3GC-R) Escherichia coli responsible for human and pet infections in Europe.
METHODS
Whole-genome sequencing data from 3GC-R E. coli isolated from clinical samples of humans, dogs and cats, and published in eight European studies were re-analyzed using bioinformatics tools. The acquired genes responsible for 3GC-R were identified. The sequence type (ST) of all genomes were assessed by multilocus sequence typing. Alpha and beta diversities were measured within and between the two populations.
RESULTS
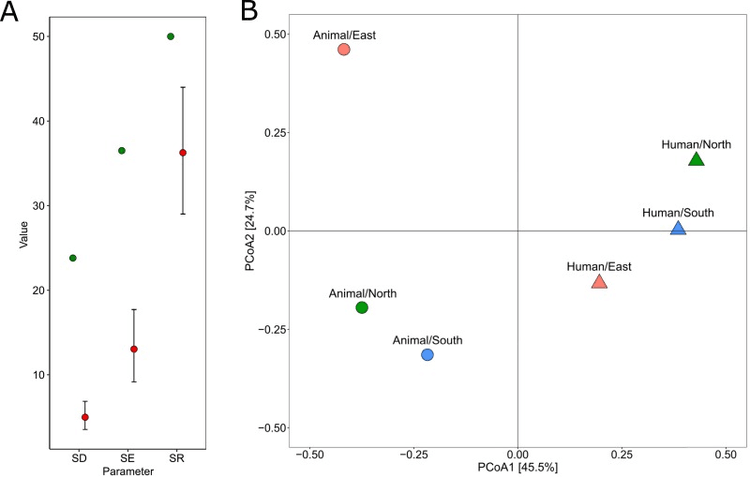
We included genomes of 1327 3GC-R E. coli isolated from humans and animals with 109 (8.2 %) being responsible for infections in dogs and cat, and 1218 (91.8 %) responsible for human infections. Alpha diversity analysis suggested greater diversity within ST and 3GC-R genes in the animal population. Beta diversity analysis by principal coordinate analysis separated animal and human strains. ST131 was more abundant in human strains (43.4 %) than in animal strains (14.7 %) (p < 0.001). Six STs, including ST372, were identified almost exclusively in 3GC-R E. coli from animal origin. The blaCTX-M-15 gene was more frequent in humans (49.24 %) than in companion animals (17.9 %) (p < 0.001). The resistance genes blaCMY-2 (30.8 %) and blaCTX-M-1 (15.4 %) were more frequent in E. coli isolated from pets (p < 0.001).
CONCLUSION
We found that populations of 3GC-R E. coli responsible for human and pet infections in Europe do not overlap. Although it cannot rule out occasional transmission of bacteria between pets and humans within a household, it suggests that dogs and cats are not a major source of human infection with this antibiotic-resistant pathogen.